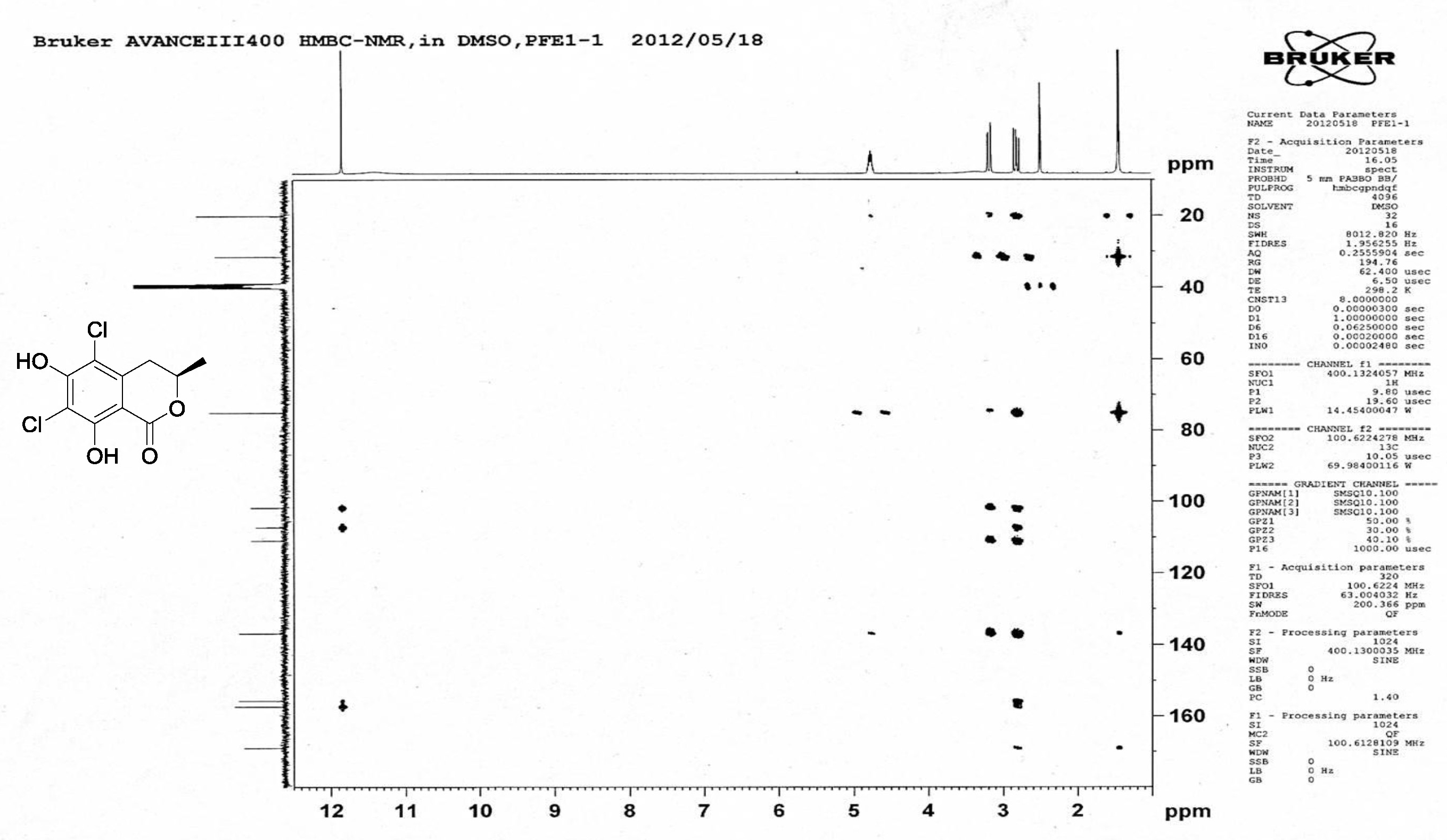
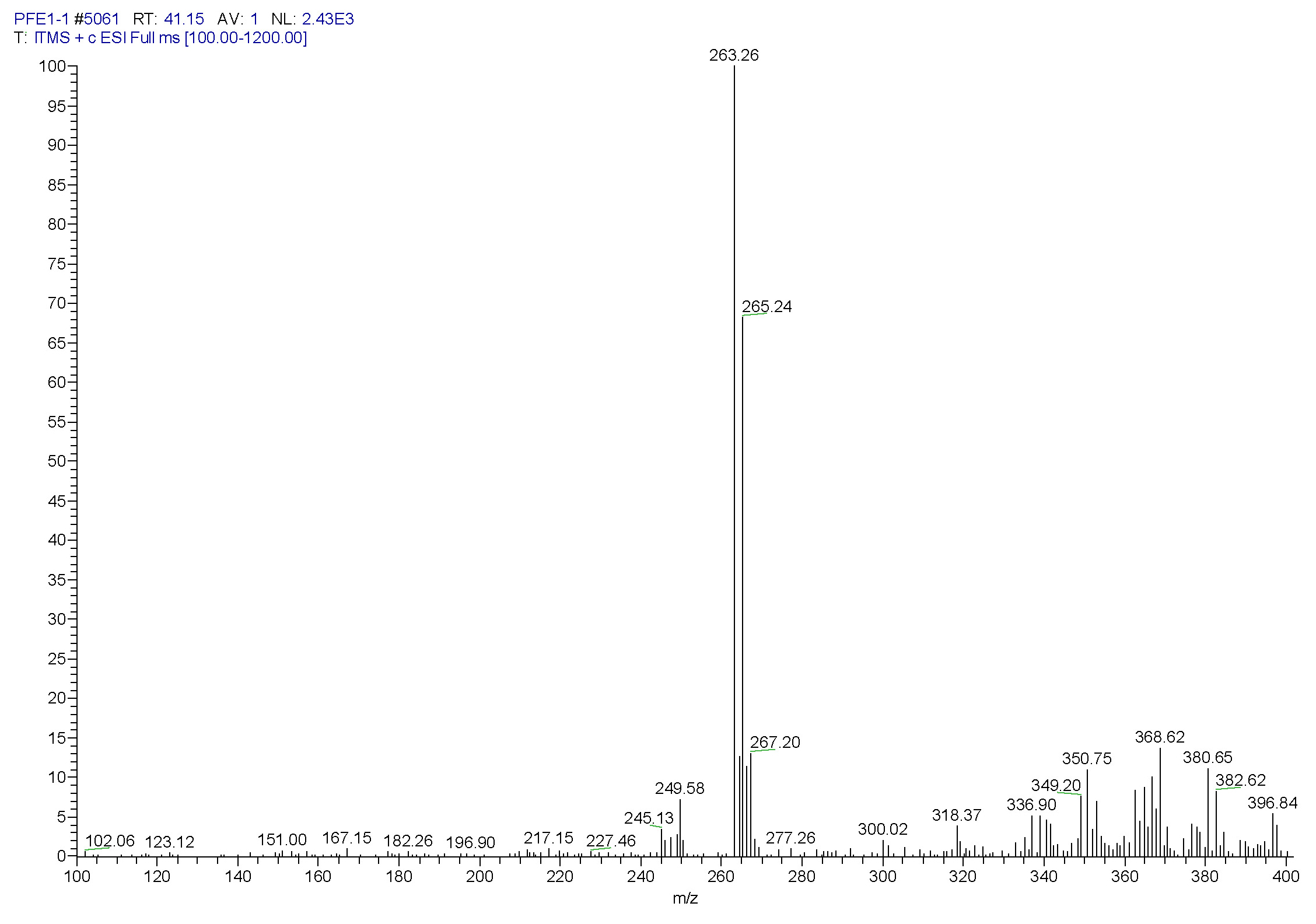
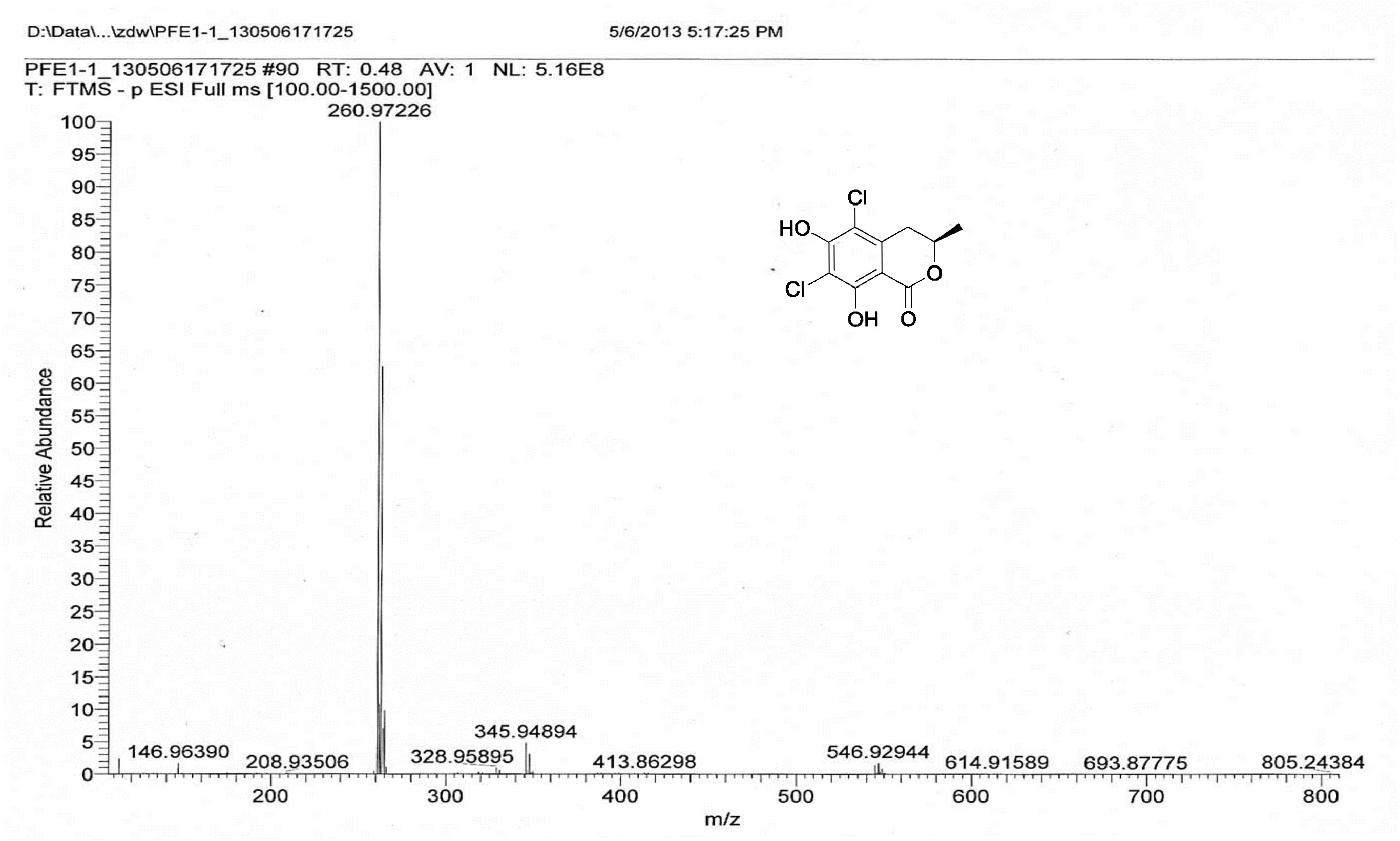
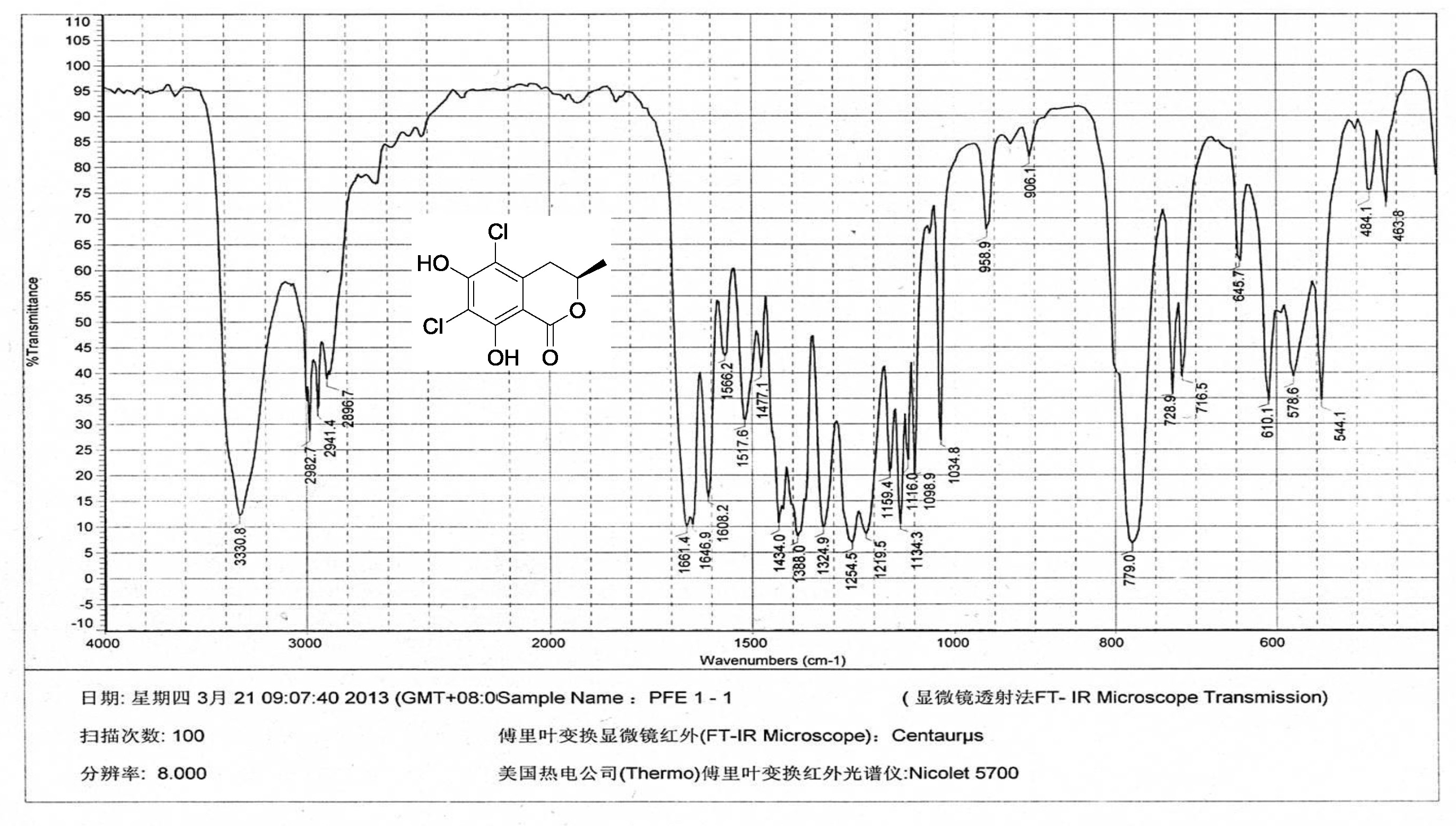
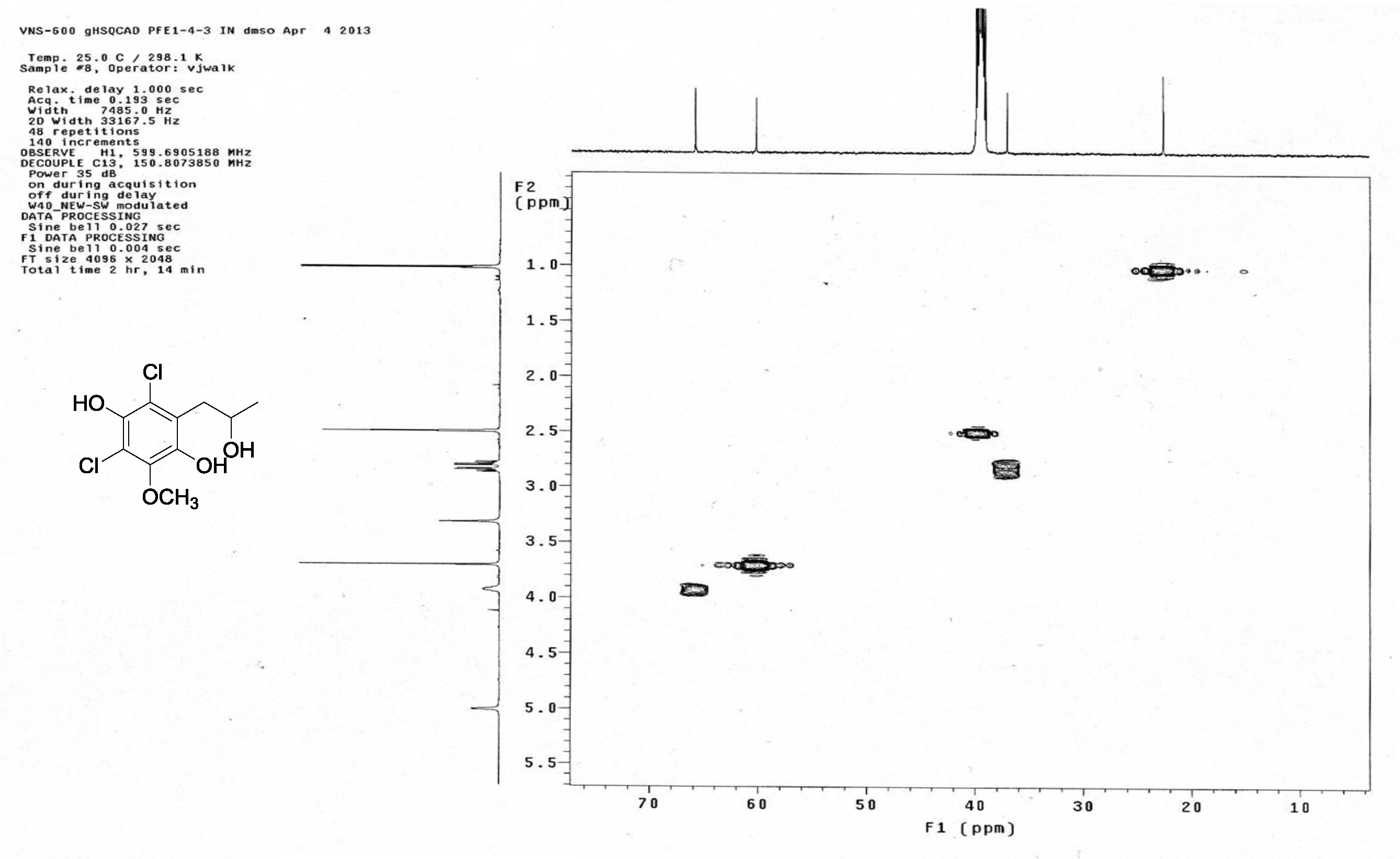
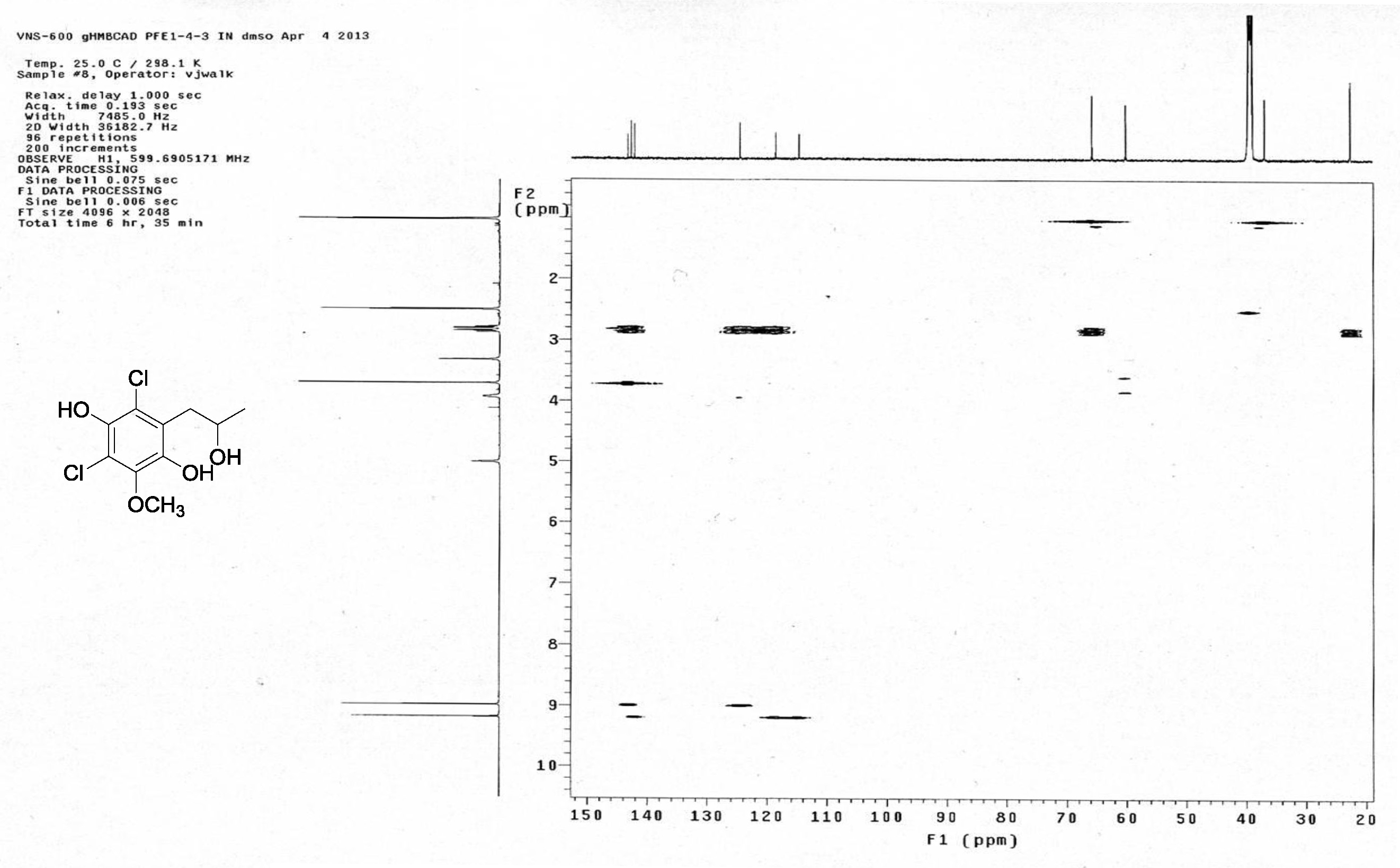
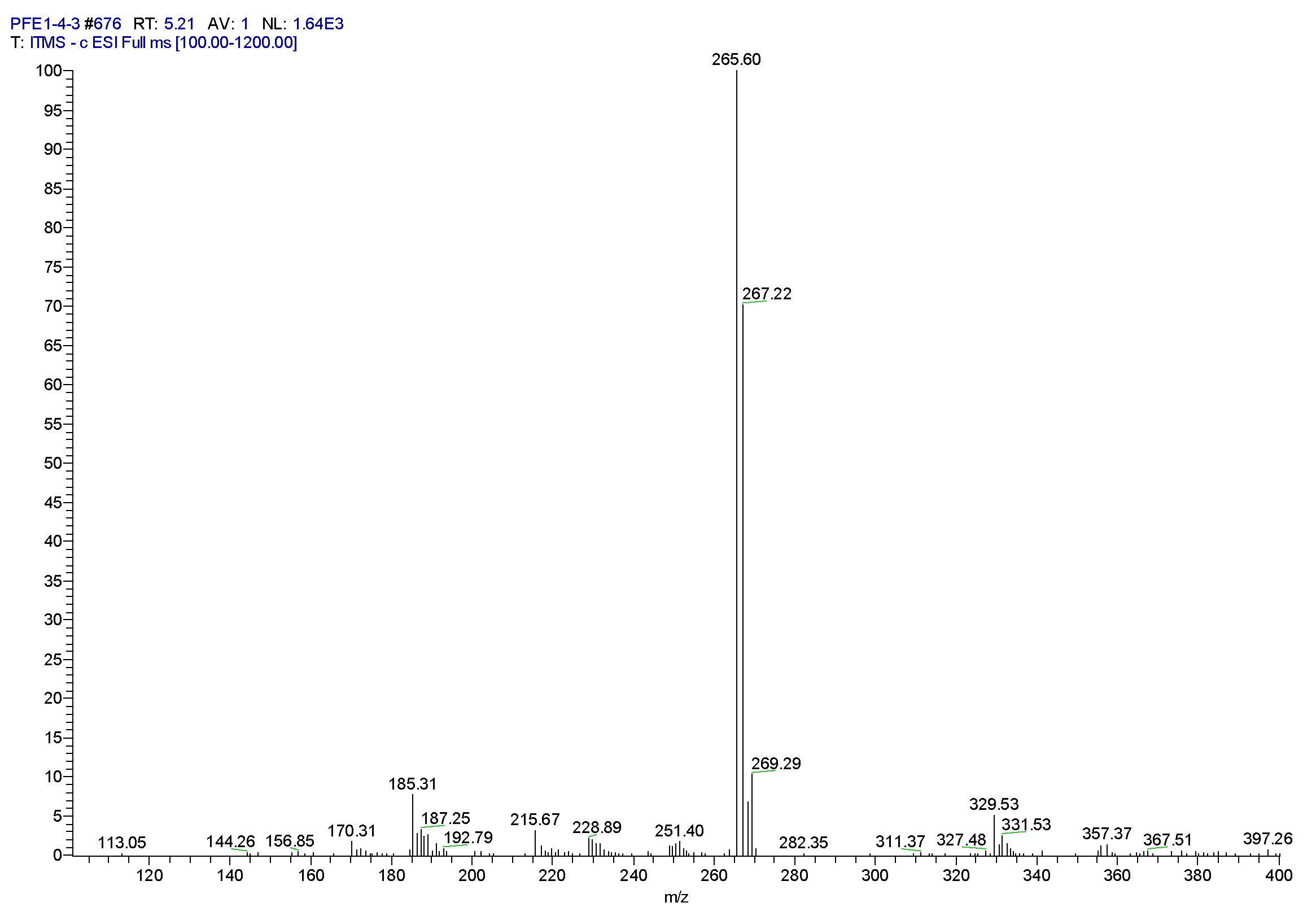
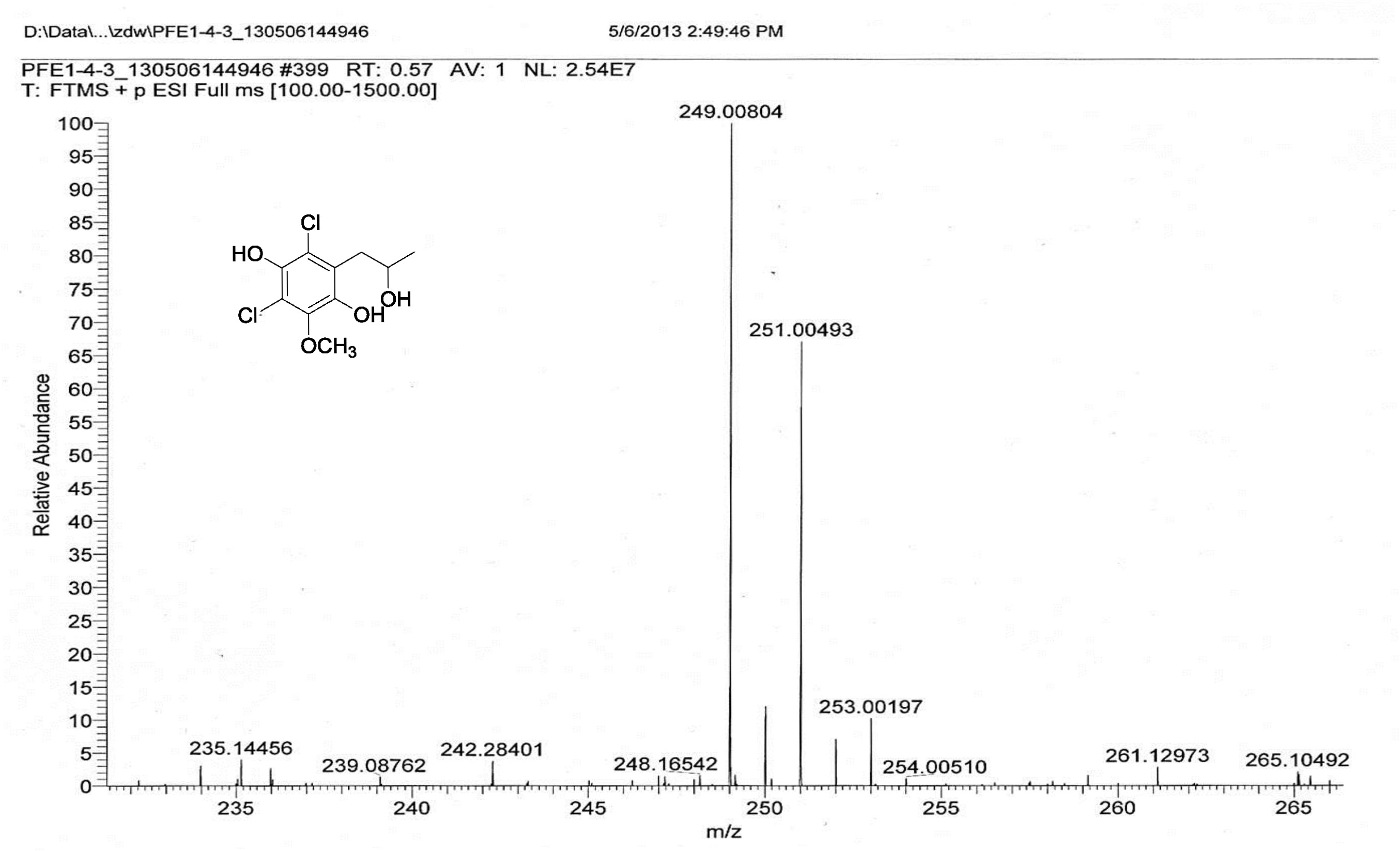
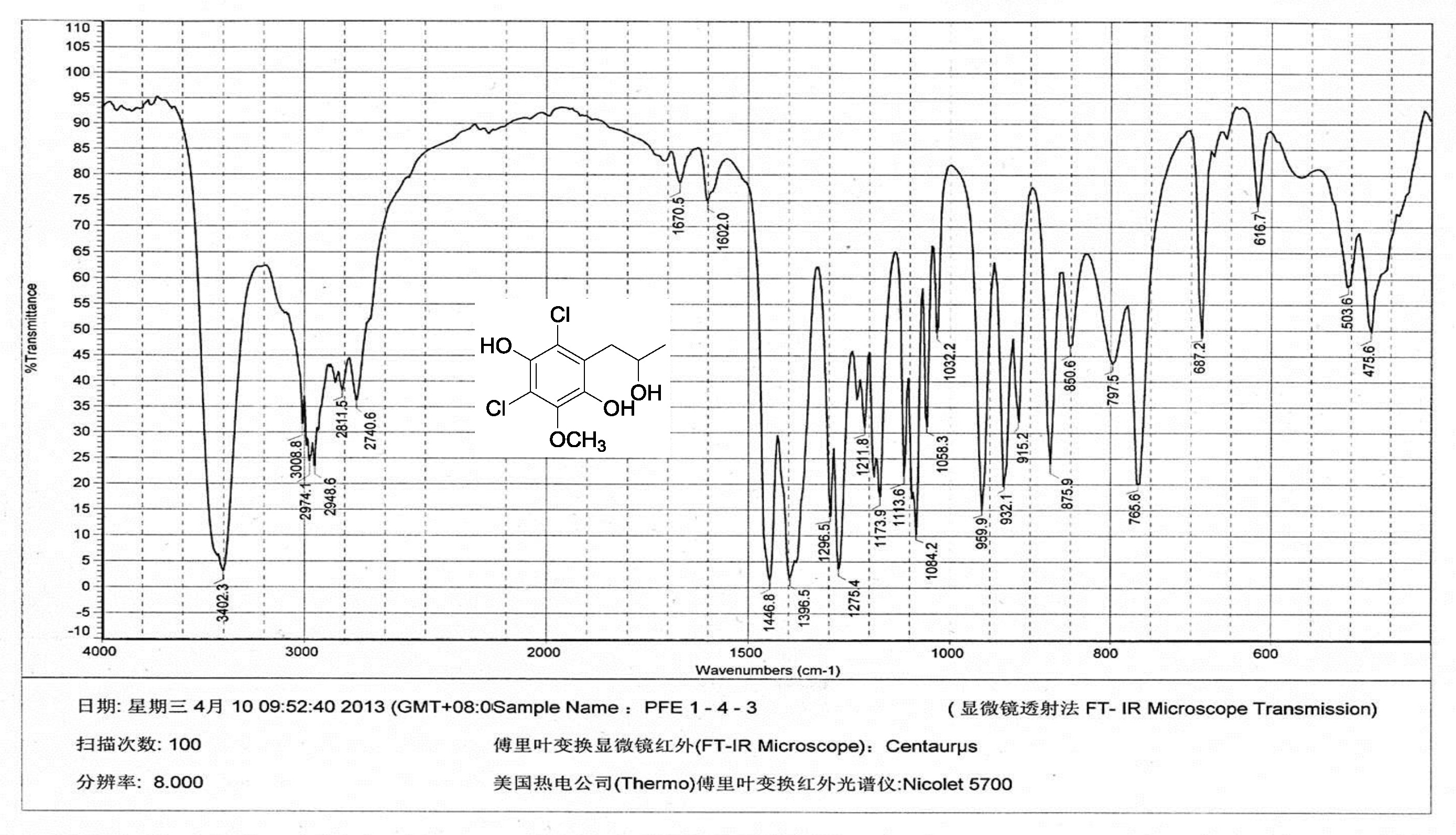
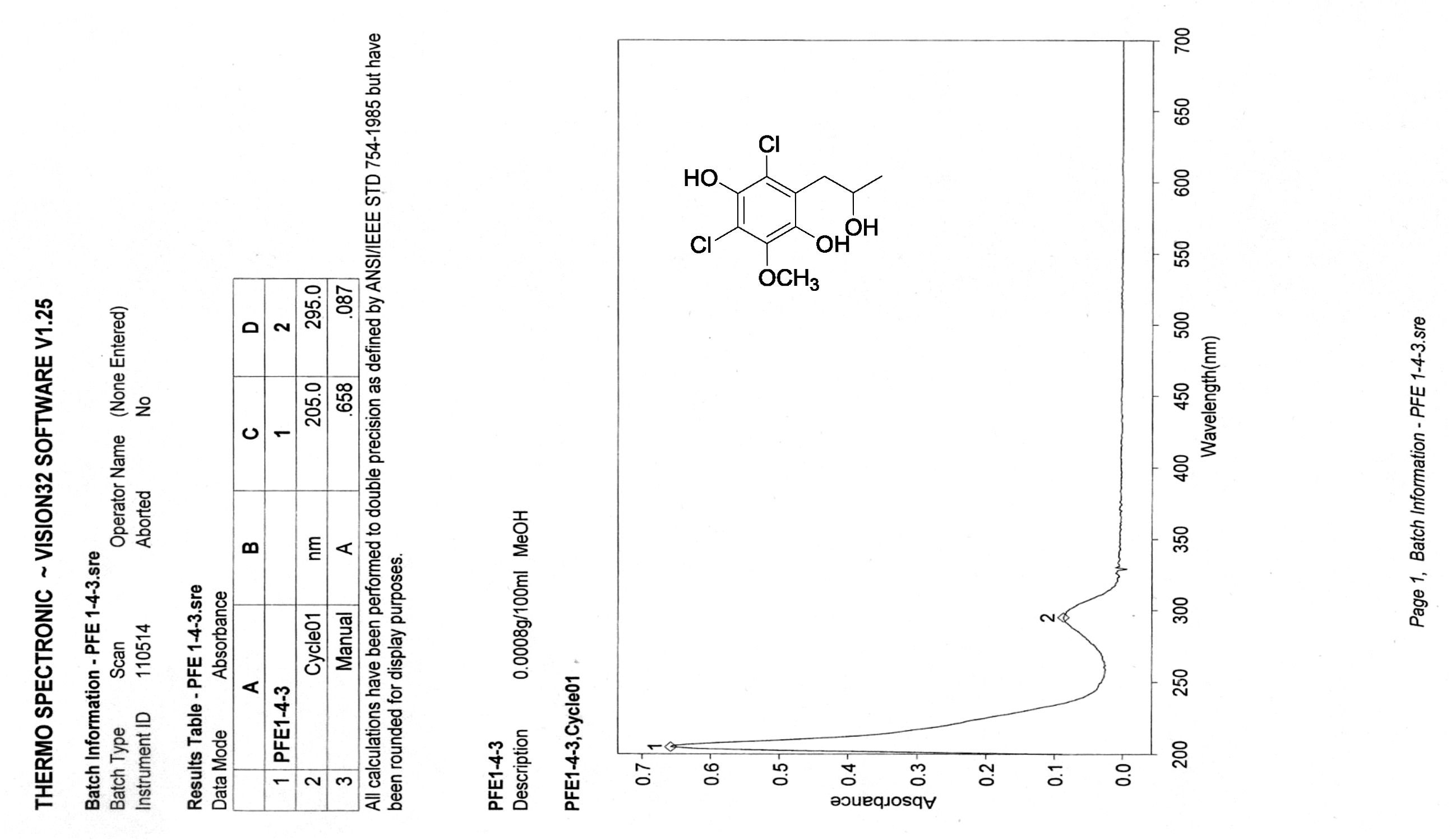
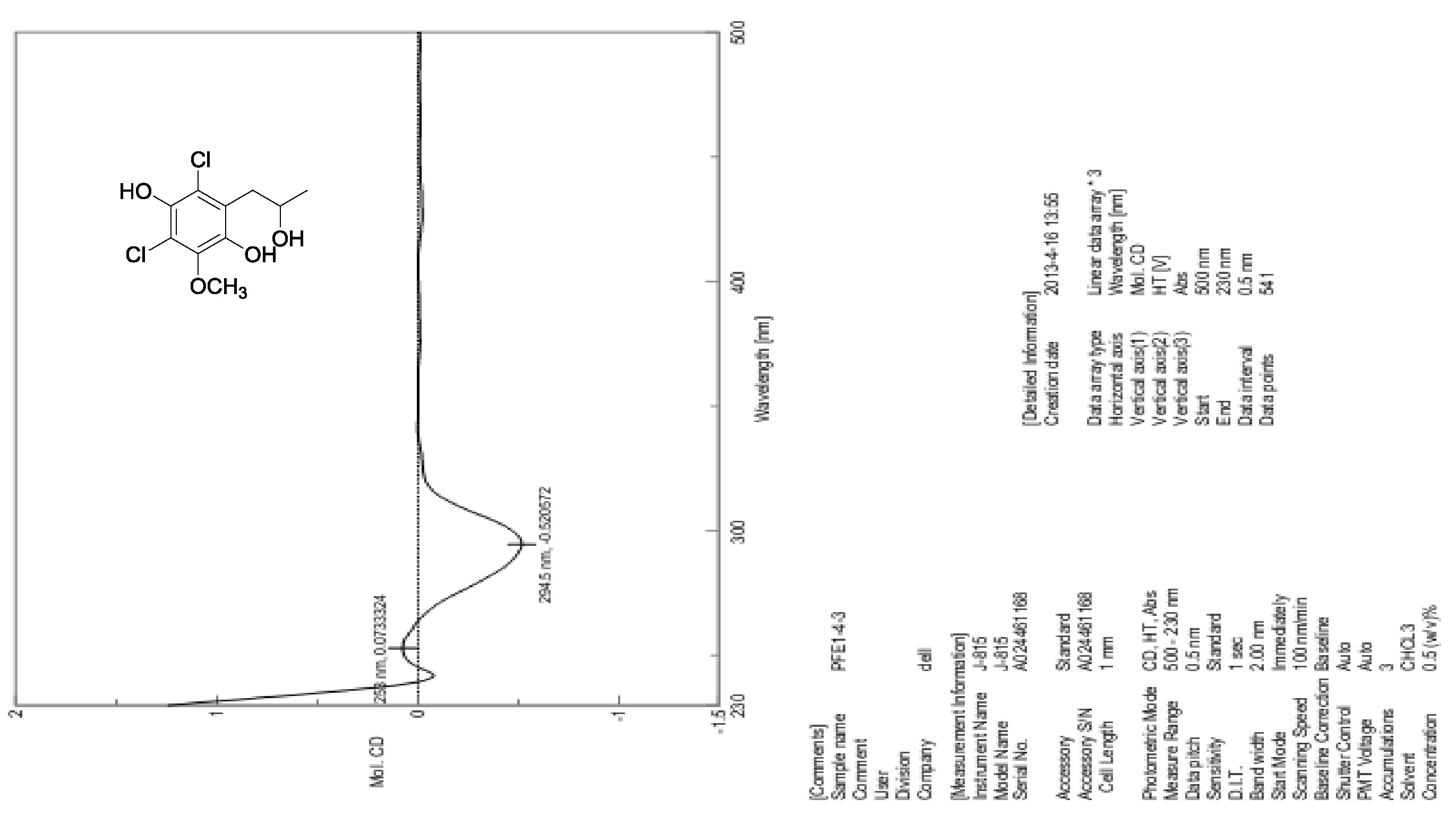
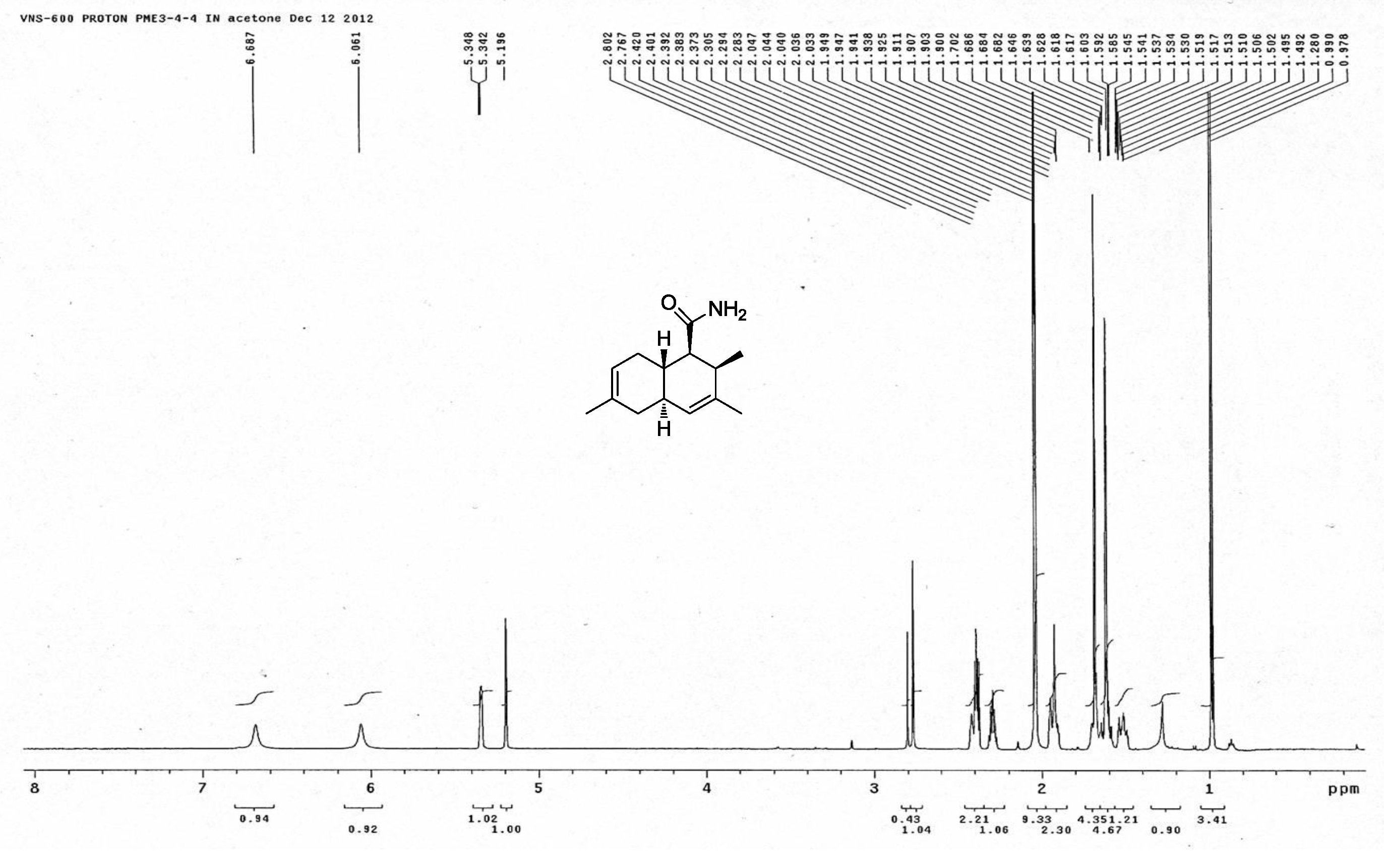
| [1] Schulz, B.; Boyle, C.; Draeger, S.; Römmert, A.K.; Krohn, K. Endophytic fungi: a source of novel biologically active secondary metabolites. Mycol. Res. 2002, 106, 996-1004.
[2] Kharwar, R.N.; Mishra, A.; Gond, S.K.; Stierle, A.; Stierle, D. Anticancer compounds derived from fungal endophytes: their importance and future challenges. Nat. Prod. Rep. 2011, 28, 1208-1228.
[3] Gubiani, J.R.; Zeraik, M.L.; Oliveira, C.M.; Ximenes, V.F.; Nogueira, C.R.; Fonseca, L.M.; Silva, D.H.; Bolzani, V.S.; Araujo, A.R. Biologically active eremophilane-type sesquiterpenes from Camarops sp., an endophytic fungus isolated from Alibertia macrophylla. J. Nat. Prod. 2014, 77, 668-672.
[4] Liu, Y.B.; Li, Y.; Qu, J.; Ma, S.G.; Zang, C.X.; Zhang, Y.T.; Yu, S.S. Eremophilane sesquiterpenes and polyketones produced by an endophytic Guignardia fungus from the toxic plant Gelsemium elegans. J. Nat. Prod. 2015, 78, 2149-2154.
[5] He, W.W.; Xu, Y.C.; Fu, P.; Zuo, M.X.; Liu, W.; Jiang, Y.M.; Wang, L.P.; Zhu, W.M. Cytotoxic indolyl diketopiperazines from the Aspergillus sp. GZWMJZ-258, endophytic with the medicinal and edible plant Garcinia multiflora. J. Agric. Food Chem. 2019, 67, 10660-10666.
[6] Wu, J.C.; Hou, Y.N.; Xu, Q.H.; Jin, X.J.; Chen, Y.X.; Fang, J.G.; Hu, B.R.; Wu, Q.X. (±)-alternamgin, a pair of enantiomeric polyketides, from the endophytic fungi Alternaria sp. MG1. Org. Lett. 2019, 21, 1551-1554.
[7] Yang, H.X.; Ai, H.L.; Feng, T.; Wang, W.X.; Wu, B.; Zheng, Y.S.; Sun, H.; He, J.; Li, Z.H.; Liu, J.K. Trichothecrotocins A-C, antiphytopathogenic agents from potato endophytic fungus Trichothecium crotocinigenum. Org. Lett. 2018, 20, 8069-8072.
[8] Zhang, D.W.; Ge, H.L.; Xie, D.; Chen, R.D.; Zou, J.H.; Tao, X.Y.; Dai, J.G. Periconiasins A-C, new cytotoxic cytochalasans with an unprecedented 9/6/5 tricyclic ring system from endophytic fungus Periconia sp. Org. Lett. 2013, 15, 1674-1677.
[9] Zhang, D.W.; Ge, H.L.; Zou, J.H.; Tao, X.Y.; Chen, R.D.; Dai, J.G. Periconianone A, a new 6/6/6 carbocyclic sesquiterpenoid from endophytic fungus Periconia sp. with neural anti-inflammatory activity. Org. Lett. 2014, 16, 1410-1413.
[10] Zhang, D.W.; Tao, X.Y.; Chen, R.D.; Liu, J.M.; Li, L.; Fang, X.M.; Yu, L.Y.; Dai, J.G. Pericoannosin A, a polyketide synthase-nonribosomal peptide synthetase hybrid metabolite with new carbon skeleton from the endophytic fungus Periconia sp. Org. Lett. 2015, 17, 4304-4307.
[11] Liu, J.M.; Zhang, D.W.; Zhang, M.; Zhao, J.L.; Chen, R.D.; Wang, N.; Zhang, D.; Dai, J.G. Eremophilane sesquiterpenes from an endophytic fungus Periconia species. J. Nat. Prod. 2016, 79, 2229-2235.
[12] Liu, J.M.; Zhang, D.W.; Zhang, M.; Liu, X.; Chen, R.D.; Zhao, J.L.; Li, Wang, N.; Dai, J.G. Periconiasins I and J, two new cytochalasans from an endophytic fungus Periconia sp. Tetrahedron Lett. 2016, 57, 5794-5797.
[13] Liu, J.M.; Zhang, D.W.; Zhang, M.; Chen, R.D.; Yan, Z.; Zhao, J.Y.; Zhao, J.L.; Wang, N.; Dai, J.G. Periconones B-E, new meroterpenoids from endophytic fungus Periconia sp. Chin. Chem. Lett. 2017, 28, 248-252.
[14] Liu, J.M.; Zhang, D.W.; Du, W.Y.; Zhang, M.; Zhao, J.L.; Chen, R.D.; Xie, K.B.; Dai, J.G. Four new monoterpenoids from an endophytic fungus Periconia sp. F-31. J. Asian. Nat. Prod. Res. 2017, 19, 541-549.
[15] Fan, Y.J.; Zhang, D.W.; Tao, X.Y.; Wang, Y.H.; Liu, J.M.; Li, L.; Zhao, J.Y.; Yu, L.Y.; He, Y.P.; Dai, J.G.; Tang, Y.F. Biosynthetic hypothesis-guided discovery and total syntheses of PKS-NRPS hybrid metabolites from endophytic fungus Periconia species. Org. Lett. 2019, 21, 1794-1798.
[16] Carmichael, J.; DeGraff, W.G.; Gazdar, A.F.; Minna, J.D.; Mitchell, J.B. Evaluation of a tetrazolium-based semiautomated colorimetric assay: assessment of chemosensitivity testing. Cancer Res. 1987, 47, 936-942.
[17] Zhang, Q.; Liu, Z.L.; Mi, Z.Y.; Li, X.Y.; Jia, P.P.; Zhou, J.M.; Yin, X.; You, X.F.; Yu, L.Y.; Guo, F.; Ma, J.; Liang, C.; Cen, S. High-throughput assay to identify inhibitors of Vpu-mediated down-regulation of cell surface BST-2. Antiviral Res. 2011, 91, 321-329.
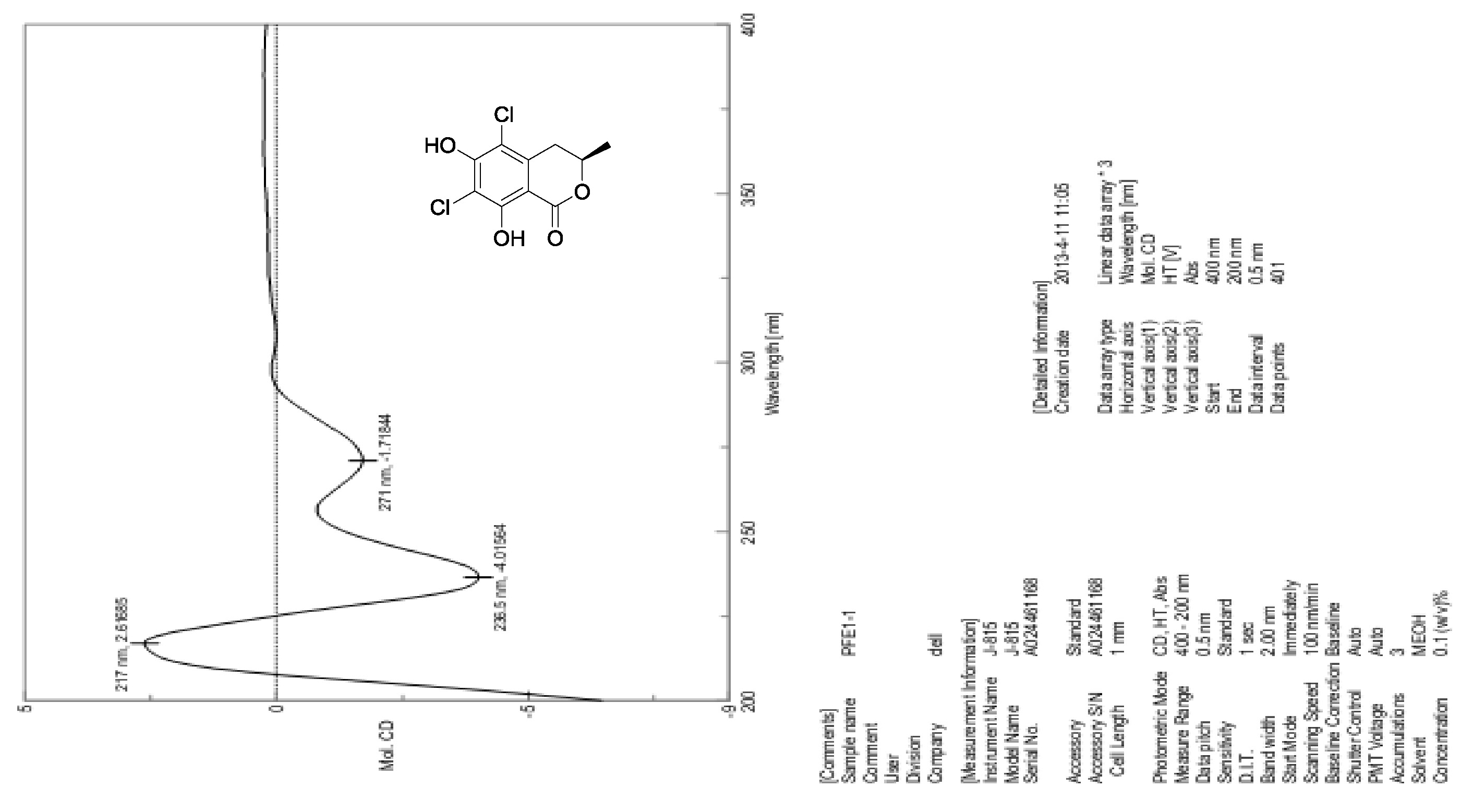
[18] Krohn, K.; Bahramsari, R.; Flörke, U.; Ludewig, K.; Kliche-Spory, C.; Michel, A.; Aust, H.J.; Draeger, S.; Schulz, B.; Antus, S. Dihydroisocoumarins from fungi: isolation, structure elucidation, circular dichroism and biological activity. Phytochemistry. 1997, 45, 313-320.
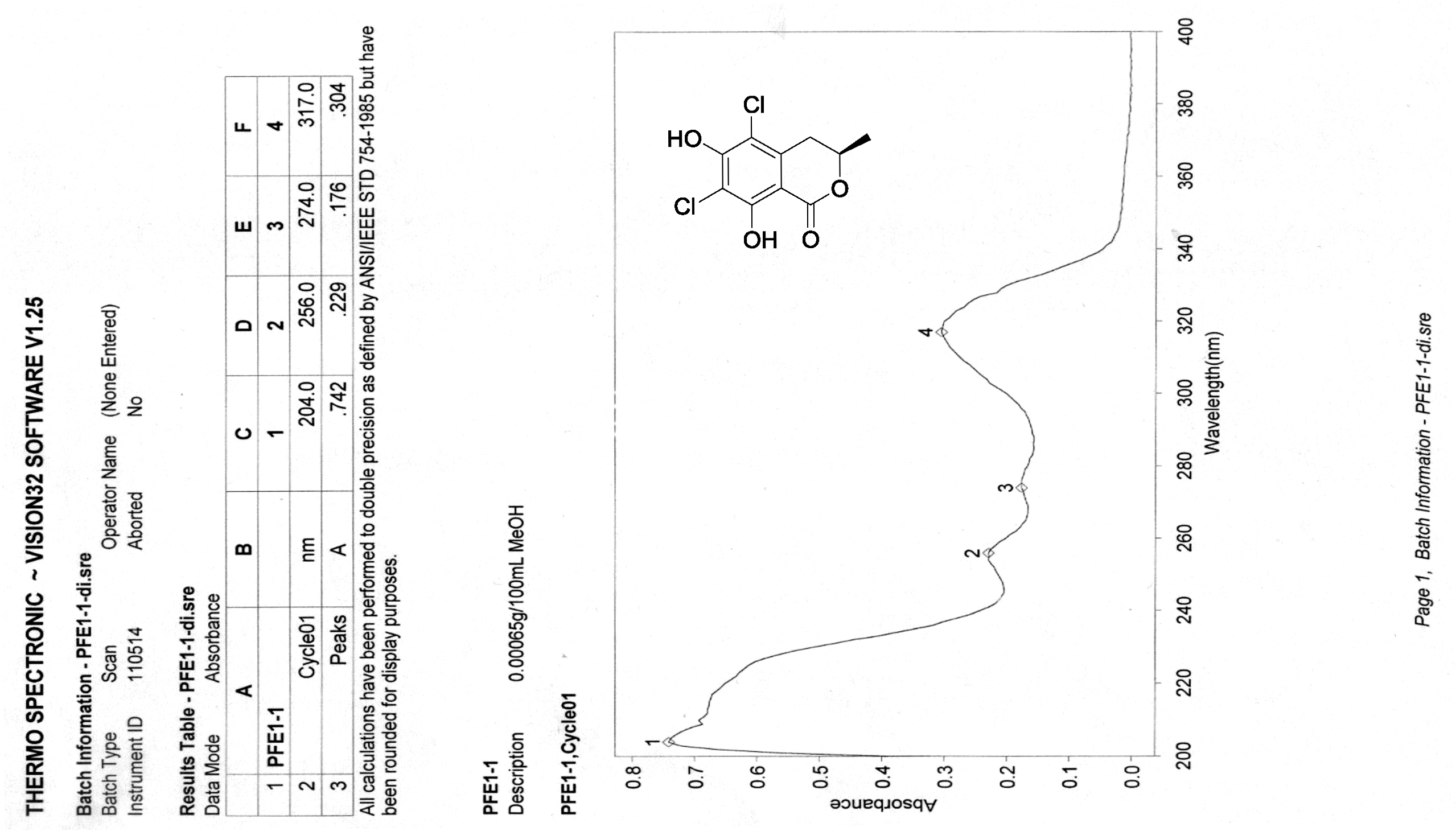
[19] Henderson, G.B.; Hill, R.A. Synthesis of chlorinated isocoumarin derivatives. J. Chem. Soc. Perkin Trans. 1982, 1, 1111-1115.
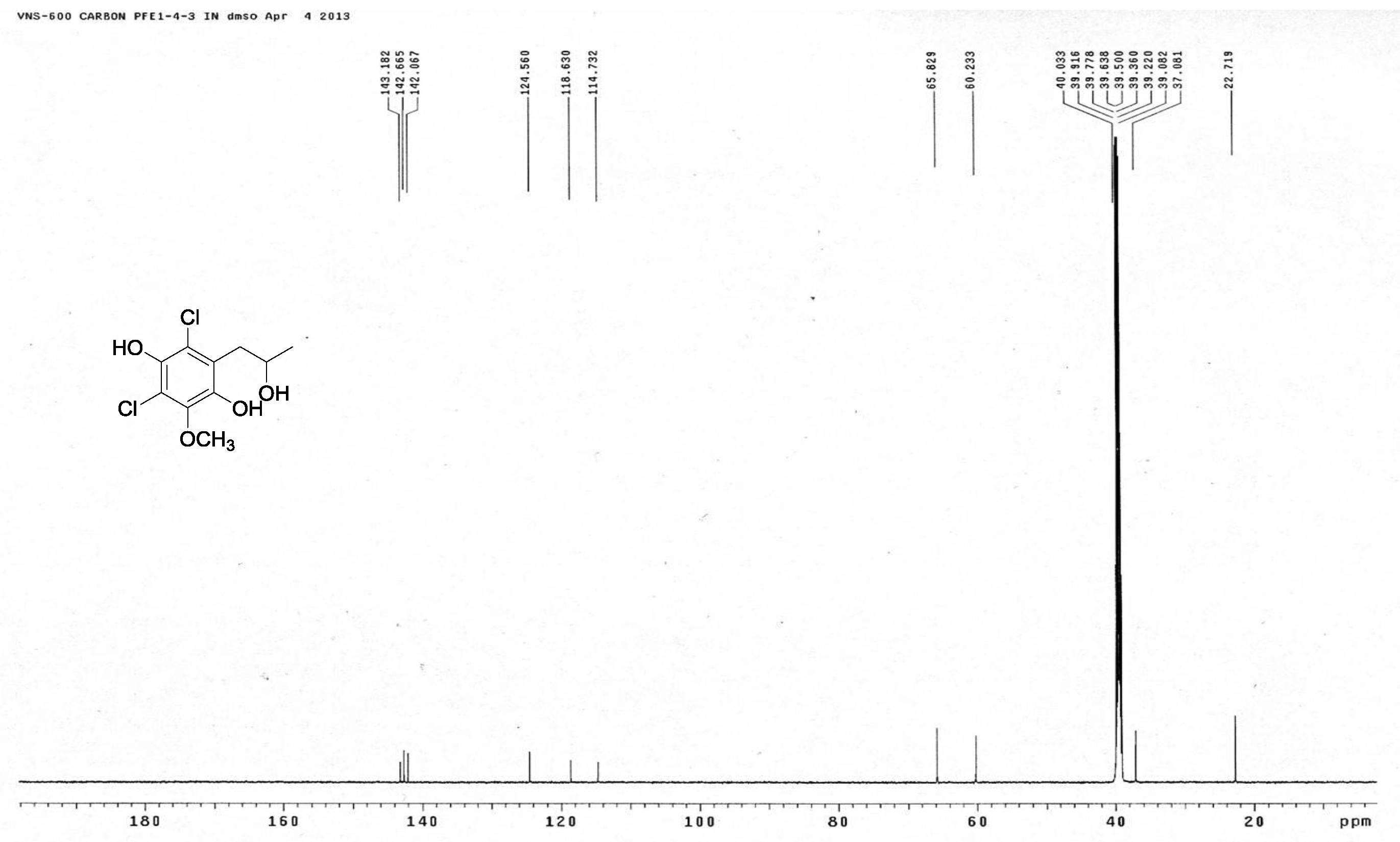
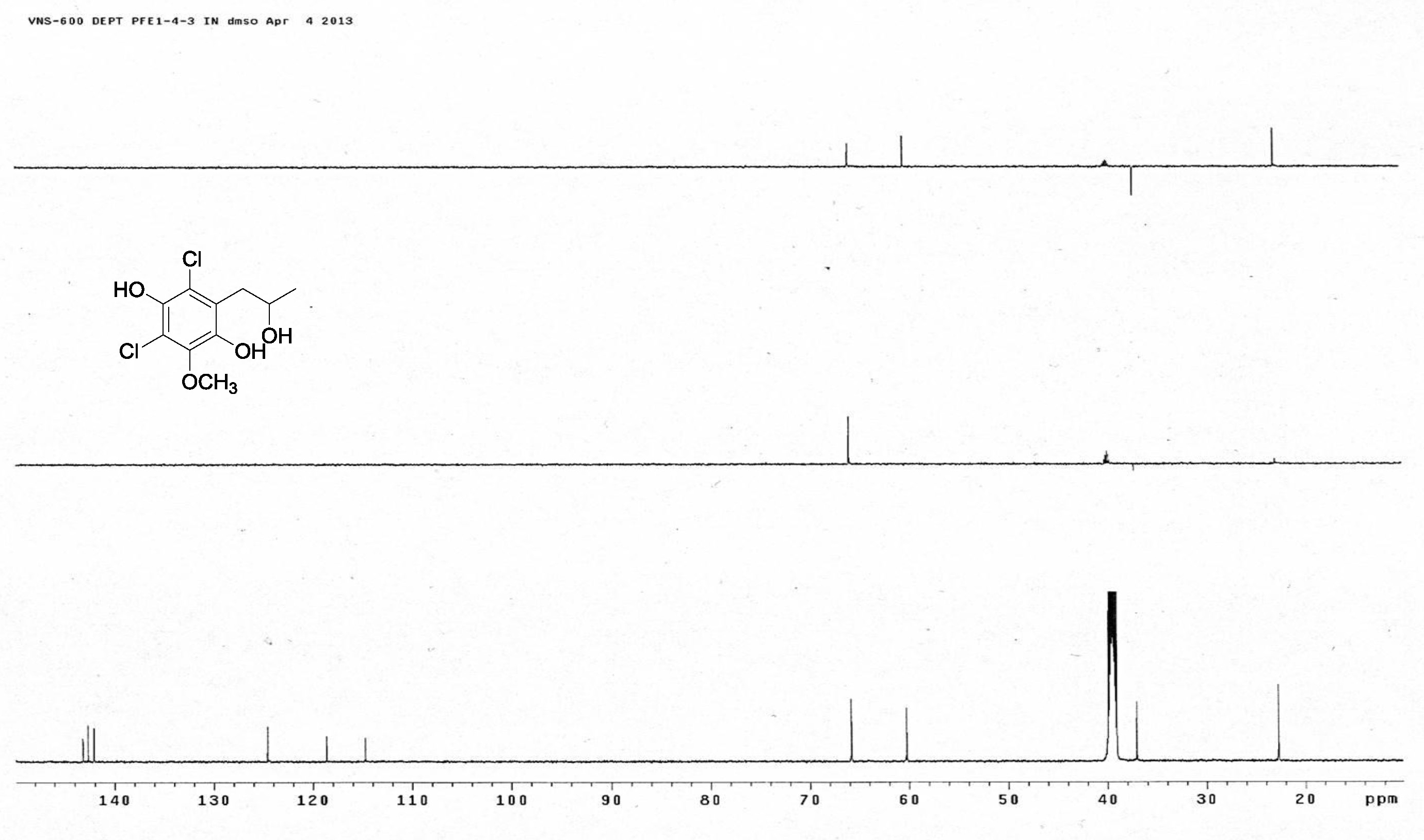
[20] Giles, D.; Turner, W.B. Chlorine-containing metabolites of Periconia macrospinosa. J. Chem. Soc. Perkin Trans. 1969, 16, 2187-2189. |
.jpg)
.jpg)
.jpg)