中国药学(英文版) ›› 2017, Vol. 26 ›› Issue (8): 545-555.DOI: 10.5246/jcps.2017.08.061
• 【研究论文】 • 下一篇
刘园, 陈亚, 胡建星, 刘振明*, 张亮仁*
Yuan Liu, Ya Chen, Jianxing Hu, Zhenming Liu*, Liangren Zhang*
摘要:
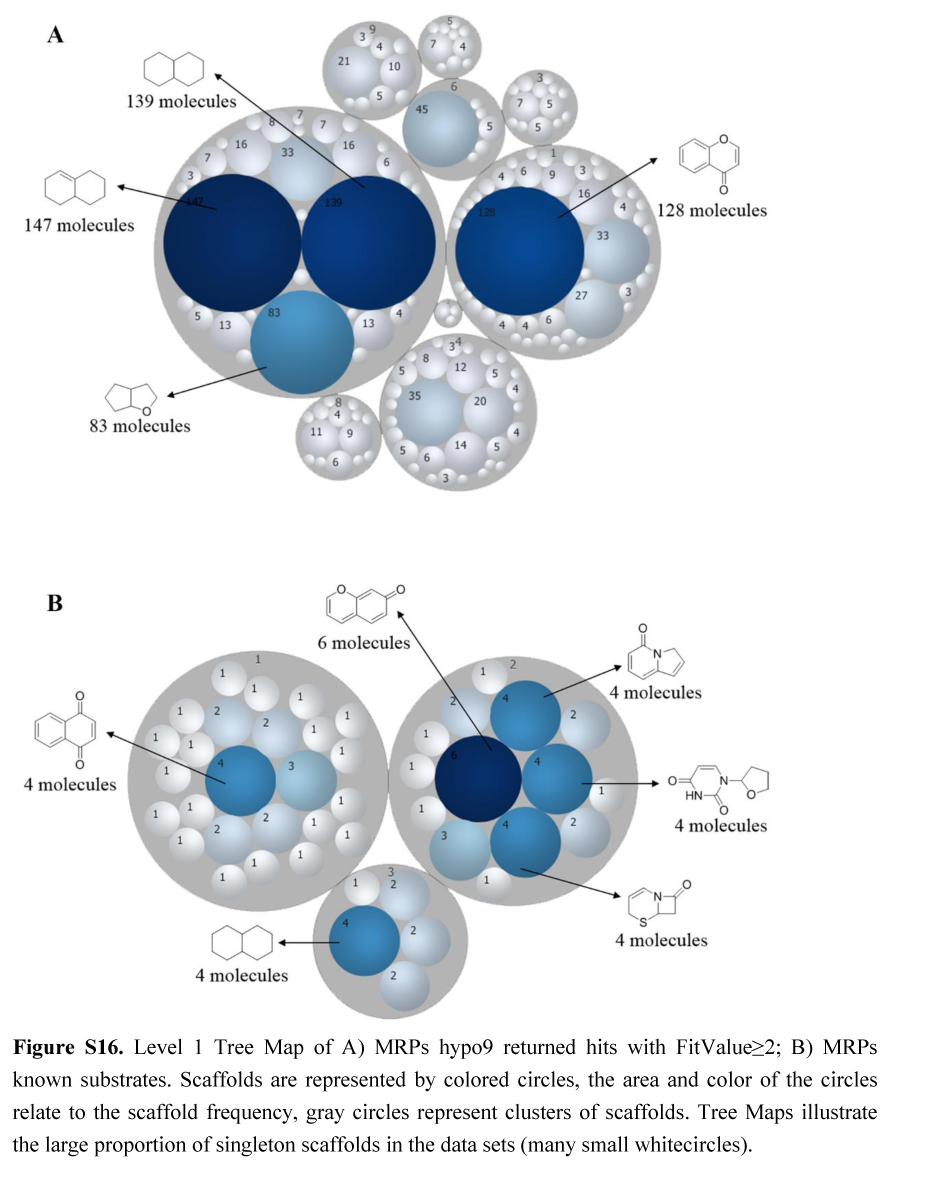
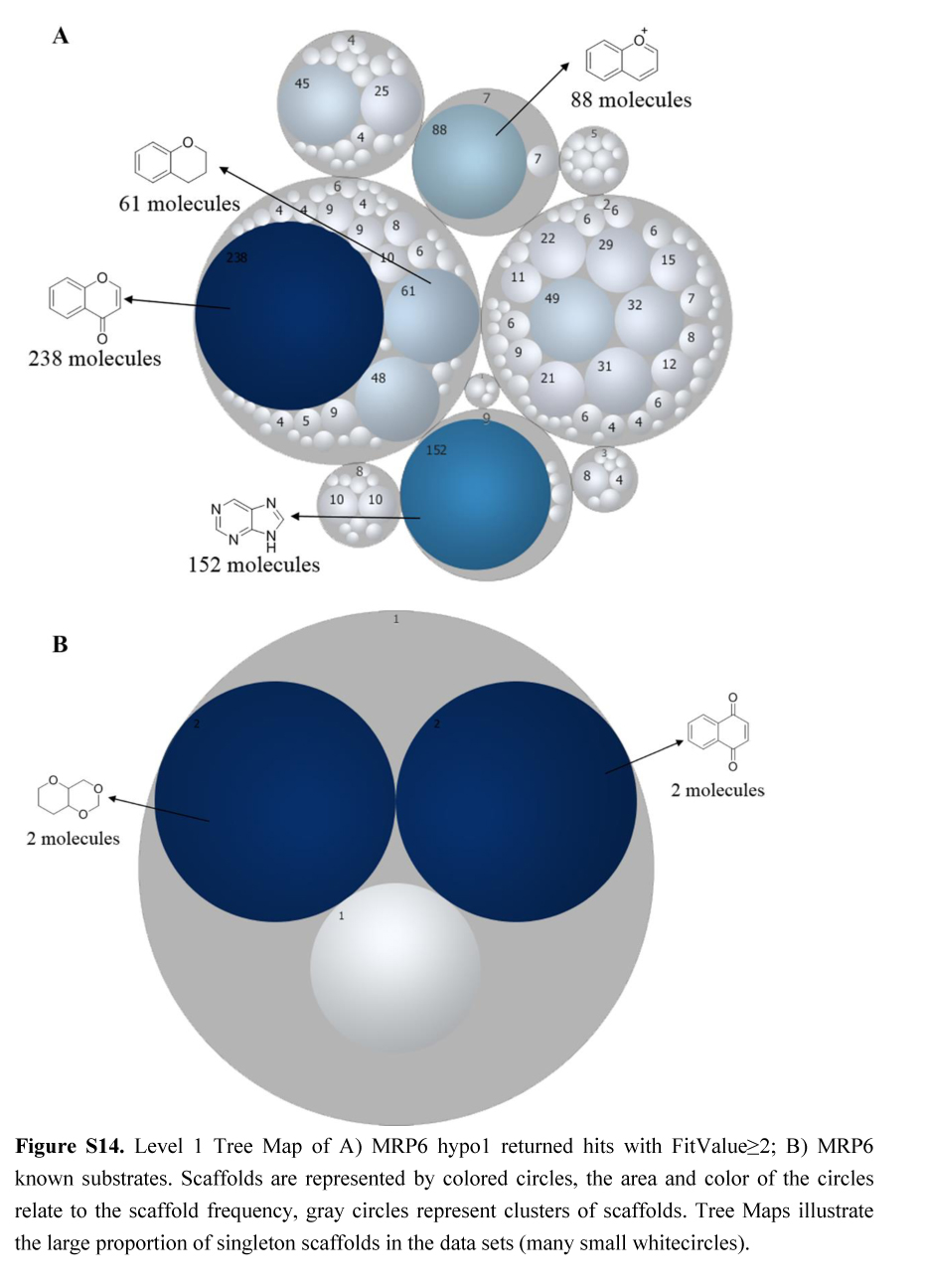
多药耐药相关蛋白(Multidrug resistance-associated proteins, MRPs)是在人体广泛分布的一类跨膜转运蛋白。MRPs可选择性和特异性地将不同结构的药物、药物结合物及代谢物和其他的小分子化合物转到细胞外,使临床应用的抗肿瘤药物产生耐药性。因此,准确预测MRPs可转运的特异性底物分子特征,对于抗肿瘤药物的抗耐药性研发具有重要意义。本文利用MRPs的7个亚型(包括MRP1, -2, -3, -4, -5, -6和-8)已知的底物分子和MRPs整个家族共同转运的底物分子,应用CATALYST软件,基于分子共同特征分别构建了MRPs各亚型和整个家族的药效团模型。并利用DUD-E生成诱饵分子用于验证和选取药效团模型,选择AUC(area under curve)打分最好的药效团模型对人体内源性代谢数据库(HMDB)进行筛选,筛选获得的多个分子得到了文献的验证。通过对药效团筛选出的分子和已知的底物分子进行物理性质(ALOGP、分子极性表面、分子体积、分子质量、氢键受体数和氢键给体数)和结构骨架的比较分析,发现两者: 1)两者在ALOGP、分子体积、分子质量上具有一致的分布趋势; 2)与其他亚型相比, MRP1的底物分子具有较高的脂溶性,这与MRP1药效团模型中具有两个疏水中心相一致; 3)两者在骨架特征上,具有相同的骨架结构或相似的骨架片段。
中图分类号:
Supporting:
|
Name
|
Training set substrates structures
|
|
MRP1
|
 |
|
MRP2
|
 |
|
MRP3
|
 |
|
MRP4
|
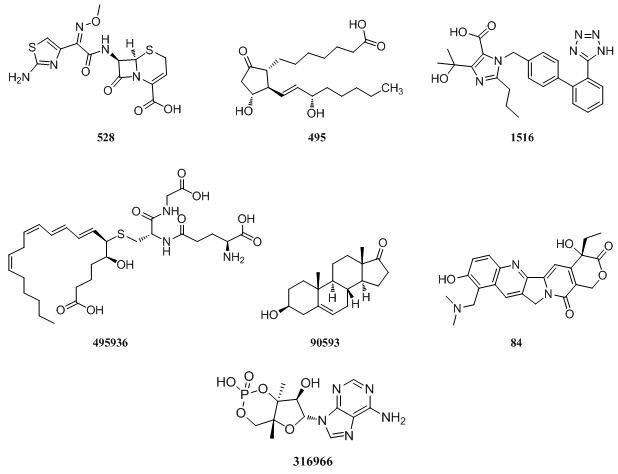 |
|
MRP5
|
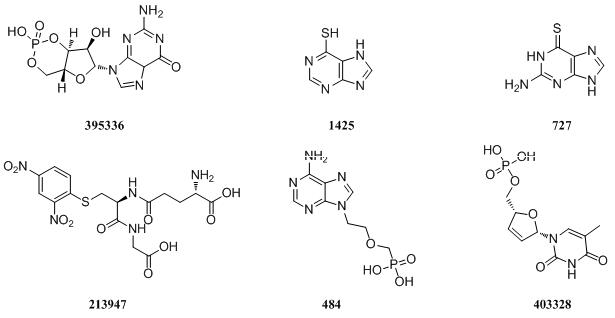 |
|
MRP6
|
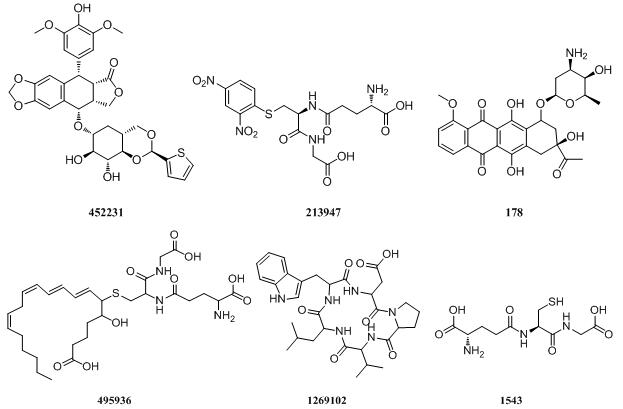 |
|
MRP8
|
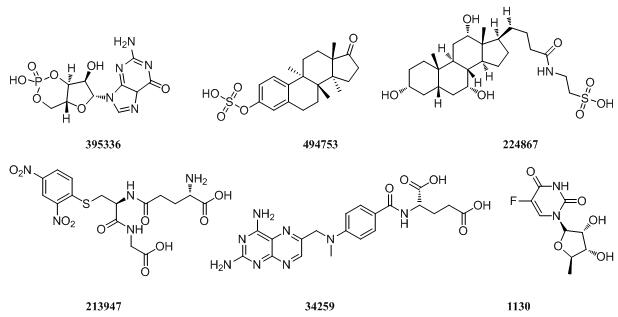 |
|
MRPs
|
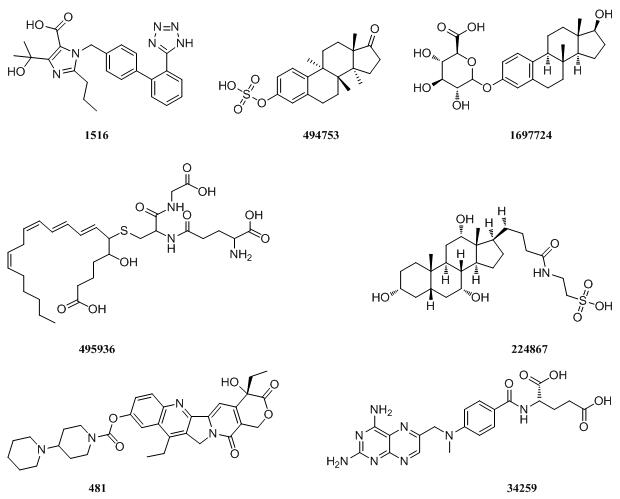 |
|
Name
|
Hypotheses rank
|
Features
|
Scores
|
|
MRP1
|
hypo1
|
HHAA
|
49.533
|
|
|
hypo2
|
HHAA
|
48.682
|
|
|
hypo3
|
HHAA
|
48.605
|
|
|
hypo4
|
HHAA
|
48.323
|
|
|
hypo5
|
HHAA
|
48.323
|
|
|
hypo6
|
HHAA
|
47.872
|
|
|
hypo7
|
HHAA
|
47.714
|
|
|
hypo8
|
HHAA
|
47.264
|
|
|
hypo9
|
HHAA
|
47.11
|
|
|
hypo10
|
HHAA
|
46.968
|
|
MRP2
|
hypo1
|
AAA
|
61.918
|
|
|
hypo2
|
AAA
|
61.918
|
|
|
hypo3
|
AAA
|
61.346
|
|
|
hypo4
|
AAA
|
61.01
|
|
|
hypo5
|
AAA
|
60.962
|
|
|
hypo6
|
AAA
|
60.767
|
|
|
hypo7
|
AAA
|
60.706
|
|
|
hypo8
|
AAA
|
60.493
|
|
|
hypo9
|
AAA
|
60.294
|
|
|
hypo10
|
AAA
|
59.89
|
|
MRP3
|
hypo1
|
DAA
|
65.895
|
|
|
hypo2
|
DAA
|
65.273
|
|
|
hypo3
|
DAA
|
65.244
|
|
|
hypo4
|
DAA
|
65.175
|
|
|
hypo5
|
AAA
|
65.175
|
|
|
hypo6
|
DAA
|
65.019
|
|
|
hypo7
|
AAA
|
64.668
|
|
|
hypo8
|
AAA
|
64.497
|
|
|
hypo9
|
DAA
|
64.445
|
|
|
hypo10
|
AAA
|
64.4
|
|
MRP4
|
hypo1
|
NDA
|
50.69
|
|
|
hypo2
|
NDA
|
50.67
|
|
|
hypo3
|
NDA
|
50.375
|
|
|
hypo4
|
NAA
|
49.335
|
|
|
hypo5
|
NDA
|
49.33
|
|
|
hypo6
|
NDA
|
49.266
|
|
|
hypo7
|
NDA
|
49.259
|
|
|
hypo8
|
NDA
|
49.255
|
|
|
hypo9
|
NDA
|
49.189
|
|
|
hypo10
|
NDA
|
49.138
|
|
MRP5
|
hypo1
|
NAAA
|
61.686
|
|
|
hypo2
|
NAAA
|
61.457
|
|
|
hypo3
|
NAAA
|
61.044
|
|
|
hypo4
|
NAAA
|
60.857
|
|
|
hypo5
|
NAAA
|
60.715
|
|
|
hypo6
|
NAAA
|
60.372
|
|
|
hypo7
|
NAAA
|
60.249
|
|
|
hypo8
|
NAAA
|
59.785
|
|
|
hypo9
|
NAAA
|
59.37
|
|
|
hypo10
|
NAAA
|
59.304
|
|
MRP6
|
hypo1
|
AAAAA
|
85.327
|
|
|
hypo2
|
AAAAA
|
85.288
|
|
|
hypo3
|
AAAAA
|
84.432
|
|
|
hypo4
|
AAAAA
|
84.426
|
|
|
hypo5
|
AAAAA
|
84.419
|
|
|
hypo6
|
AAAAA
|
84.361
|
|
|
hypo7
|
AAAAA
|
84.274
|
|
|
hypo8
|
AAAAA
|
84.17
|
|
|
hypo9
|
AAAAA
|
84.159
|
|
|
hypo10
|
AAAAA
|
84.046
|
|
MRP8
|
hypo1
|
NAA
|
51.598
|
|
|
hypo2
|
NAA
|
51.359
|
|
|
hypo3
|
NAA
|
50.389
|
|
|
hypo4
|
NAA
|
50.314
|
|
|
hypo5
|
NAA
|
49.878
|
|
|
hypo6
|
NAA
|
49.47
|
|
|
hypo7
|
NAA
|
49.409
|
|
|
hypo8
|
NAA
|
49.409
|
|
|
hypo9
|
NAA
|
49.465
|
|
|
hypo10
|
NAA
|
49.089
|
|
MRPs
|
hypo1
|
HHAAA
|
91.794
|
|
|
hypo2
|
HHAAA
|
91.794
|
|
|
hypo3
|
HHAAA
|
91.756
|
|
|
hypo4
|
HHAAA
|
91.671
|
|
|
hypo5
|
HHAAA
|
91.516
|
|
|
hypo6
|
HHAAA
|
91.458
|
|
|
hypo7
|
HHAAA
|
91.182
|
|
|
hypo8
|
HHAAA
|
91.182
|
|
|
hypo9
|
HHAAA
|
91.182
|
|
|
hypo10
|
HHAAA
|
91.173
|
.jpg)
.jpg)
.jpg)
.jpg)
.jpg)
.jpg)
.jpg)
.jpg)
.jpg)
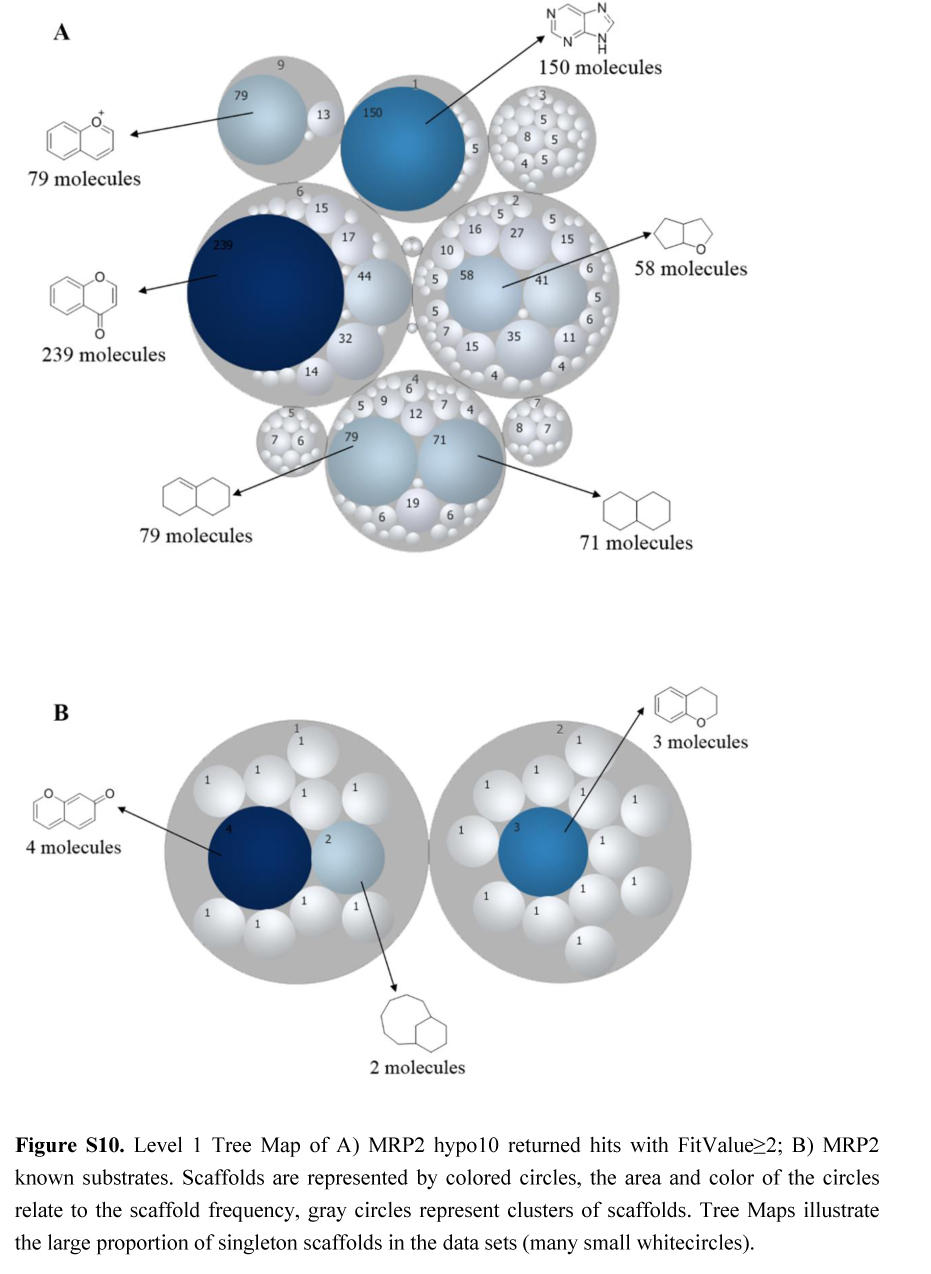
.jpg)
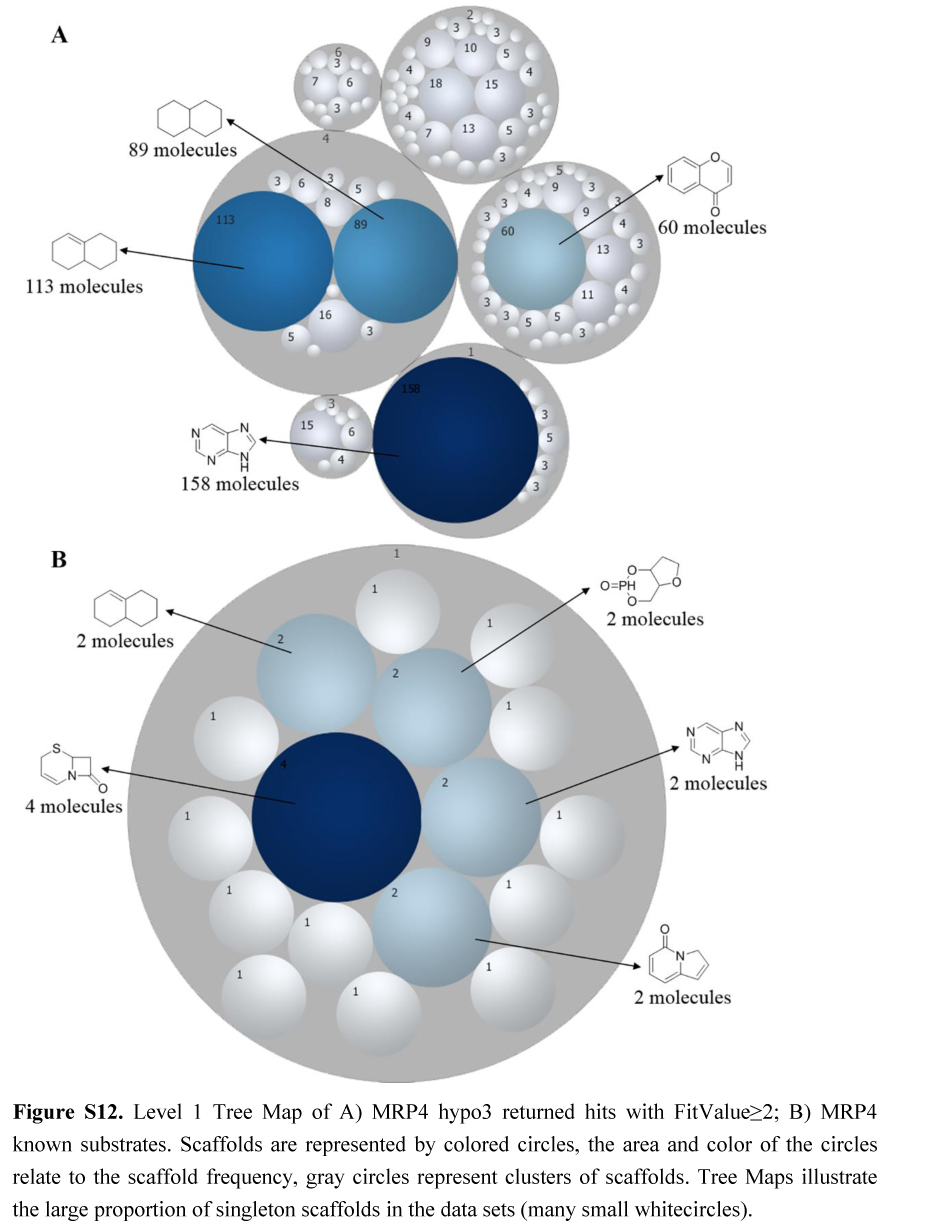
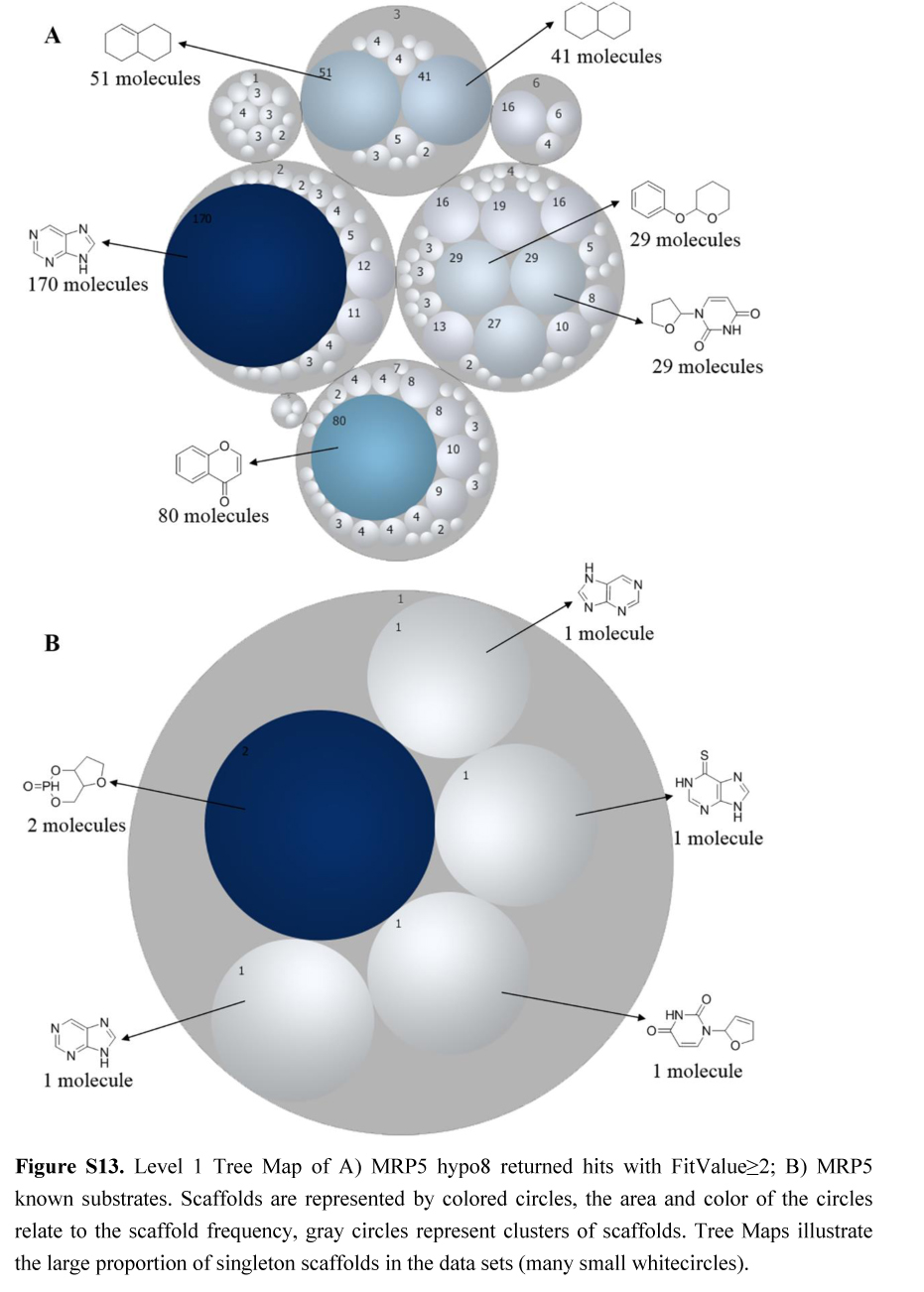
.jpg)